About me
I’m an assistant professor in Institute of Science Tokyo. I have proposed several methods with computational, algorithmical, and physicochemical consideration.
Recent news
2025 July-December
- 2025/12 A poster presentation will be given at The international Chemical Congress of Pacific Basin Societies (Pacifichem) 2025.
- Chie Motono, Keisuke Yanagisawa, Jun Koseki, Kenichiro Imai. “CrypTothML: Cryptic Site Prediction using Mixed-Solvent Molecular Dynamics Simulation and Machine Learning”, The international Chemical Congress of Pacific Basin Societies (Pacifichem) 2025, 2025/12/15-20.
- 2025/10 Poster presentations will be given at Chem-Bio Informatics Society(CBI) Annual Meeting 2025.
- Keisuke Yanagisawa, Ryunosuke Yoshino, Genki Kudo, Takatsugu Hirokawa. “Quantitative Estimation of Protein-Ligand Substructure Interaction with Inverse Mixed-Solvent Molecular Dynamics Simulation”, Chem-Bio Informatics Society (CBI) Annual Meeting 2025, P01-26, 2025/10/27-30.
- Masatake Sugita, Kei Terakura, Takuya Fujie, Keisuke Yanagisawa, Yutaka Akiyama. “Analysis of membrane permeation processes of cyclic peptides on multiple reaction coordinates based on the Markov state model”, Chem-Bio Informatics Society (CBI) Annual Meeting 2025, P01-06, 2025/10/27-30.
- Kaho Akaki, Keisuke Yanagisawa, Yutaka Akiyama. “Enhancing virtual screening accuracy by refining docking calculation scoring with mixed-solvent molecular dynamics”, Chem-Bio Informatics Society (CBI) Annual Meeting 2025, P06-16, 2025/10/27-30.
- Masahiro Shimizu, Masatake Sugita, Keisuke Yanagisawa, Yutaka Akiyama. “Development of an automatic parameter adjustment method for REST/REUS MD and its application to predicting the membrane permeability of cyclic peptides”, Chem-Bio Informatics Society (CBI) Annual Meeting 2025, P01-17, 2025/10/27-30.
- Masayoshi Shimizu, Keisuke Yanagisawa, Yutaka Akiyama. “COFFEE-PRESC: a fast pre-screening method using chemical compound retrieval by fragment pose pairs”, Chem-Bio Informatics Society (CBI) Annual Meeting 2025, P06-13, 2025/10/27-30.
- Ryoya Nakano, Keisuke Yanagisawa, Yutaka Akiyama. “Improvement of fragment-based protein–ligand docking using the Quantum Annealer”, Chem-Bio Informatics Society (CBI) Annual Meeting 2025, P06-19, 2025/10/27-30.
- Satoshi Yoneyama, Keisuke Yanagisawa, Yutaka Akiyama. “Construction of representative fragment sets based on mutual 3D structural similarity and docking feasibility for fragment-based virtual screening”, Chem-Bio Informatics Society (CBI) Annual Meeting 2025, P06-15, 2025/10/27-30.
- 2023/09 A poster presentation will be given at the 63rd Annual Meeting of the Biophysical Society of Japan.
- Masatake Sugita, Kei Terakura, Takuya Fujie, Keisuke Yanagisawa, Yutaka Akiyama. “Kinetic analysis of membrane permeation processes of cyclic peptides on multiple reaction coordinates based on the Markov state model”, The 63rd Annual Meeting of The Biophysical Society of Japan, 2Pos175, 2025/09/24-26.
- 2025/09 Invited oral presentations will be given at Asia Hub for e-Drug Discovery (AHeDD) 2025. Five poster presentations will be also given.
- Keisuke Yanagisawa, Takuya Fujie, Kazuki Takabatake, Yutaka Akiyama. “FraSCO-VS: Fragment-based drug virtual screening by combinatorial optimization with quantum annealer”, Asia Hub for e-Drug Discovery 2025 (AHeDD2025), 2025/9/24. (invited talk)
- Keisuke Yanagisawa, Ryunosuke Yoshino, Genki Kudo, Takatsugu Hirokawa. “Quantitative Evaluation of Protein-Ligand Substructure Interaction with Inverse Mixed-Solvent Molecular Dynamics Simulation”, Asia Hub for e-Drug Discovery 2025 (AHeDD2025), 2025/9/24.
- Masatake Sugita, Yudai Noso, Jianan Li, Takuya Fujie, Keisuke Yanagisawa, Yutaka Akiyama. “Protocol for Membrane Permeability Prediction of Cyclic Peptides by Combining Molecular Dynamics Simulations and Machine Learning”, Asia Hub for e-Drug Discovery 2025 (AHeDD2025), 2025/9/24.
- Kaho Akaki, Keisuke Yanagisawa, Yutaka Akiyama. “Enhancing virtual screening accuracy by refining docking calculation scoring with mixed-solvent molecular dynamics”, Asia Hub for e-Drug Discovery 2025 (AHeDD2025), 2025/9/24.
- Masahiro Shimizu, Masatake Sugita, Keisuke Yanagisawa, Yutaka Akiyama. “An Automatic Iterative Refinement Protocol for Restraint Parameters in REUS Molecular Dynamics”, Asia Hub for e-Drug Discovery 2025 (AHeDD2025), 2025/9/24.
- Masayoshi Shimizu, Satoshi Yoneyama, Keisuke Yanagisawa, Yutaka Akiyama. “Development of a fast pre-screening method using compound retrieval by fragment pose pairs”, Asia Hub for e-Drug Discovery 2025 (AHeDD2025), 2025/9/24.
2025 January-June
- 2025/06 Oral presentations will be given at IPSJ SIG Bioinformatics and Genomics (SIGBIO).
- Ryoya Nakano, Keisuke Yanagisawa, Yutaka Akiyama. “Application of the Ising model to fragment-based protein-ligand docking”, IPSJ SIG Technical Report, 2025-BIO-82(46): 1-8, 2025/06/22.
- Masahiro Shimizu, Masatake Sugita, Keisuke Yanagisawa, Yutaka Akiyama. “Development of a replica parameter optimization method for REUS MD and its application to membrane permeability prediction of cyclic peptides”, IPSJ SIG Technical Report, 2025-BIO-82(45): 1-8, 2025/06/22.
- Satoshi Yoneyama, Keisuke Yanagisawa, Yutaka Akiyama. “Selection of Representative Fragment Sets for Fragment-Based Virtual Screening Focusing on 3D Structural Similarity”, IPSJ SIG Technical Report, 2025-BIO-82(44): 1-8, 2025/06/22.
- Kaho Akaki, Keisuke Yanagisawa, Yutaka Akiyama. “Probe atom distributions obtained from mixed-solvent molecular dynamics improves scoring of docking calculations”, IPSJ SIG Technical Report, 2025-BIO-82(43): 1-8, 2025/06/22.
- 2025/06 Poster presentations will be given at the 25th Annual Meeting of the Protein Science Society of Japan.
- Kaho Akaki, Keisuke Yanagisawa, Yutaka Akiyama. “Probe atoms distribution in mixed-solvent molecular dynamics improves scoring of docking calculation”, The 25th Annual Meeting of the Protein Science Society of Japan, 1P-061, 2025/06/18-20.
- Chie Motono, Keisuke Yanagisawa, Jun Koseki, Kenichiro Imai. “CrypTothML: A Combined Mixed-Solvent Molecular Dynamics Simulation and Machine Learning Method for Cryptic Site Prediction”, The 25th Annual Meeting of the Protein Science Society of Japan, 3P-038, 2025/06/18-20.
- Masahiro Shimizu, Masatake Sugita, Keisuke Yanagisawa, Yutaka Akiyama. “Development of an Optimization Method for REUS MD Parameters and Its Application to the Prediction of Cyclic Peptide Membrane Permeability”, The 25th Annual Meeting of the Protein Science Society of Japan, 1P-064, 2025/06/18-20.
- Kei Terakura, Masatake Sugita, Takuya Fujie, Keisuke Yanagisawa, Yutaka Akiyama. “Kinetic Analysis of Membrane Permeation Process of Cyclic Peptides Using Markov State Models with Molecular Dynamics Simulations”, The 25th Annual Meeting of the Protein Science Society of Japan, 2P-044, 2025/06/18-20.
- Masatake Sugita, Yudai Noso, Jianan Li, Takuya Fujie, Keisuke Yanagisawa, Yutaka Akiyama. “Protocol for Membrane Permeability Prediction of Cyclic Peptides by Combining Molecular Dynamics Simulations and Machine Learning”, The 25th Annual Meeting of the Protein Science Society of Japan, 3P-040, 2025/06/18-20.
- 2025/03 An oral presentation will be given at IPSJ SIG Bioinformatics and Genomics (SIGBIO).
- Tomoya Saito, Masayoshi Shimizu, Keisuke Yanagisawa, Yutaka Akiyama. “フラグメントの類似性を考慮した化合物立体配座検索システムの構築”, IPSJ SIG Technical Report, 2025-BIO-81(12): 1-8, 2025/03/07.
2024 July-December
- 2024/12 A poster presentation will be given at the 52nd SAR symposium.
- Genki Kudo, Keisuke Yanagisawa, Ryunosuke Yoshino, Takatsugu Hirokawa. “3D Protein-Protein Interaction Surface Profile using Mixed-Solvent Molecular Dynamics”, 第52回構造活性相関シンポジウム, KP20, 2024/12/12-13.
- 2024/10 I got a funding from Kayamori Foundation of Informational Science Advancement.
- “共溶媒分子動力学法によるタンパク質化合物ドッキング計算のスコア関数の改善”, Research Funding Program, Kayamori Foundation of Informational Science Advancement, PI, 1 000 000 yen, 2024/10-2026/09.
- 2024/10 Poster presentations will be given at Chem-Bio Informatics Society(CBI) Annual Meeting 2024.
- Tomoya Saito, Keisuke Yanagisawa, Yutaka Akiyama. “Development of an efficient compound 3D conformer search system based on relative position of fragments”, Chem-Bio Informatics Society(CBI) Annual Meeting 2024, P07-33, 2024/10/28-31.
- Masayoshi Shimizu, Keisuke Yanagisawa, Yutaka Akiyama. “Development of a compound pre-screening method based on docking of fragments”, Chem-Bio Informatics Society(CBI) Annual Meeting 2024, P07-16, 2024/10/28-31.
- Kaho Akaki, Keisuke Yanagisawa, Yutaka Akiyama. “Acquisition of Bias Information for Protein-Ligand Docking by Mixed-Solvent Molecular Dynamics”, Chem-Bio Informatics Society(CBI) Annual Meeting 2024, P07-15, 2024/10/28-31.
- Winner of the Like! Poster Award (Selected by Conference Attendee Votes)
- Keisuke Yanagisawa, Takuya Fujie, Kazuki Takabatake, Yutaka Akiyama. “QUBO Problem Formulation of Fragment-Based Protein–Compound Flexible Docking”, Chem-Bio Informatics Society(CBI) Annual Meeting 2024, P07-14, 2024/10/28-31.
- Masatake Sugita, Yudai Noso, Takuya Fujie, Jianan Li, Keisuke Yanagisawa, Yutaka Akiyama. “Development of Prediction Models for Membrane Permeability of Cyclic Peptides using 3D Descriptors obtained from Molecular Dynamics Simulations and 2D Descriptors”, Chem-Bio Informatics Society(CBI) Annual Meeting 2024, P07-05, 2024/10/28-31.
- Jun Koseki, Chie Motono, Keisuke Yanagisawa, Ryunosuke Yoshino, Takatsugu Hirokawa, Kenichiro Imai. “Development of the Cryptic Site searching method with Mixed-solvent molecular dynamics and Topological data analyses methods”, Chem-Bio Informatics Society(CBI) Annual Meeting 2024, P04-03, 2024/10/28-31.
- Jianan Li, Keisuke Yanagisawa, Yutaka Akiyama. “CycPeptMP: Development of Membrane Permeability Prediction Model of Cyclic Peptides with Multi-Level Molecular Features and Data Augmentation”, Chem-Bio Informatics Society(CBI) Annual Meeting 2024, P03-11, 2024/10/28-31.
- Kei Terakura, Masatake Sugita, Keisuke Yanagisawa, Yutaka Akiyama. “Kinetic Analysis of Membrane Permeation Process of Cyclic Peptides Using Markov State Models with Molecular Dynamics Simulations”, Chem-Bio Informatics Society(CBI) Annual Meeting 2024, P01-12, 2024/10/28-31.
- 2024/10 Poster presentations will be given at Asia & Pacific Bioinformatics Joint Conference 2024.
- Chie Motono, Jun Koseki, Keisuke Yanagisawa, Genki Kudo, Ryunosuke Yoshino, Takatsugu Hirokawa, Kenichiro Imai. “Cryptic site detection using machine learning based on mixed-solvent molecular dynamics simulations results”, Asia & Pacific Bioinformatics Joint Conference 2024, 2024/10-22-25.
- Jun Koseki, Chie Motono, Keisuke Yanagisawa, Genki Kudo, Ryunosuke Yoshino, Takatsugu Hirokawa, Kenichiro Imai. “Development of the Cryptic Site searching method with Mixed-solvent molecular dynamics and Topological data analyses methods”, Asia & Pacific Bioinformatics Joint Conference 2024, 2024/10-22-25.
- 2024/08 A paper has been published.
- Jianan Li, Keisuke Yanagisawa, Yutaka Akiyama. “CycPeptMP: enhancing membrane permeability prediction of cyclic peptides with multi-level molecular features and data augmentation”, Briefings in Bioinformatics, 25: bbae417, 2024/08. DOI: 10.1093/bib/bbae417
2024 January-June
- 2024/06 Poster presentations will be given at the 21st IUPAB and 62nd BSJ joint congress 2024.
- Keisuke Yanagisawa, Ryunosuke Yoshino, Genki Kudo, Takatsugu Hirokawa. “Quantitative Evaluation of Protein-Compound Substructure Interaction with Inverse Mixed-Solvent Molecular Dynamics Simulation”, 21st IUPAB and 62nd BSJ joint congress 2024, 28P-189, 2024/6/28.
- Masatake Sugita, Takuya Fujie, Yudai Noso, Keisuke Yanagisawa, Masahito Ohue, Yutaka Akiyama. “Development and Application of a Protocol for Predicting Membrane Permeability of Cyclic Peptides Based on Molecular Dynamics Simulations”, 21st IUPAB and 62nd BSJ joint congress 2024, 26P-210, 2024/06/26.
- 2024/06 Two oral presentations will be given at IPSJ SIG Bioinformatics and Genomics (SIGBIO).
- Masayoshi Shimizu, Keisuke Yanagisawa, Yutaka Akiyama. “Development of a compound pre-screening method based on spatial arrangement of promising fragment pairs.”, IPSJ SIG Technical Report, 2024-BIO-78(38): 1-8, 2024/06/21.
- Kaho Akaki, Keisuke Yanagisawa, Yutaka Akiyama. “Acquisition of Bias Information for Protein-Ligand Docking by Mixed-Solvent Molecular Dynamics”, IPSJ SIG Technical Report, 2024-BIO-78(37): 1-8, 2024/06/21.
- 2024/06 An oral presentation and a poster presentation will be given at the 24th Annual Meeting of the Protein Science Society of Japan.
- Keisuke Yanagisawa, Ryunosuke Yoshino, Genki Kudo, Takatsugu Hirokawa. “Quantitative Estimation of Protein-Compound Substructure Interaction with Inverse Mixed-Solvent Molecular Dynamics Simulation”, The 24th Annual Meeting of the Protein Science Society of Japan: [WS-14] Where Bioinformatics and Agrochemistry Meet, 2024/06/13.
- Masatake Sugita, Takuya Fujie, Yudai Noso, Keisuke Yanagisawa, Masahito Ohue, Yutaka Akiyama. “Development and Application of a Protocol for Predicting Membrane Permeability of Cyclic Peptides Based on Molecular Dynamics Simulations”, The 24th Annual Meeting of the Protein Science Society of Japan: [WS-3] The Present and Future of Peptide Design, 2024/06/11.
- Chie Motono, Keisuke Yanagisawa, Genki Kudo, Takatsugu Hirokawa, Kenichiro Imai. “Cryptic site prediction using mixed-solvent molecular dynamics simulation”, The 24th Annual Meeting of the Protein Science Society of Japan, 3P-063, 2024/06/11-13.
- 2024/04 A paper has been published.
- Keisuke Yanagisawa†, Takuya Fujie, Kazuki Takabatake, Yutaka Akiyama†. “QUBO Problem Formulation of Fragment-Based Protein-Ligand Flexible Docking”, Entropy, 26: 397, 2024/04. DOI: 10.3390/e26050397
- 2024/03 Three oral presentations will be given at IPSJ SIG Bioinformatics and Genomics (SIGBIO).
- Ayako Nunobe, Keisuke Yanagisawa, Yutaka Akiyama. “フラグメントに基づくバーチャルスクリーニングへの利用などを目指したフラグメント集合の選定”, IPSJ SIG Technical Report, 2024-BIO-77(31): 1-7, 2024/03/08.
- Yudai Noso, Masatake Sugita, Takuya Fujie, Keisuke Yanagisawa, Yutaka Akiyama. “分子動力学シミュレーション軌跡データから抽出した位置依存特徴量を活用した環状ペプチドの膜透過性予測”, IPSJ SIG Technical Report, 2024-BIO-77(16): 1-8, 2024/03/07.
- Jianan Li, Keisuke Yanagisawa, Yutaka Akiyama. “CycPeptMP: Development of Membrane Permeability Prediction of Cyclic Peptides with Multi-Level Molecular Features and Data Augmentation”, IPSJ SIG Technical Report, 2024-BIO-77(15): 1-8, 2024/03/07.
-
2023 July-December
- 2023/12 A paper has been published.
- Genki Kudo†, Keisuke Yanagisawa†‡, Ryunosuke Yoshino‡, Takatsugu Hirokawa. “AAp-MSMD: Amino Acid Preference Mapping on Protein-Protein Interaction Surfaces Using Mixed-Solvent Molecular Dynamics”, Journal of Chemical Information and Modeling, 63: 7768-7777, 2023/12. DOI: 10.1021/acs.jcim.3c01677
- 2023/11 A poster presentation will be given at the 61st Annual Meeting of the Biophysical Society of Japan.
- Keisuke Yanagisawa, Ryunosuke Yoshino, Genki Kudo, Takatsugu Hirokawa. “Quantitative Evaluation of Protein-Chemical Substructure Interaction with Inverse Mixed-Solvent Molecular Dynamics Simulation”, The 61st Annual Meeting of The Biophysical Society of Japan, 2Pos013, 2023/11/14-16.
- 2023/10 A poster presentation will be given at Chem-Bio Informatics Society(CBI) Annual Meeting 2023.
- Keisuke Yanagisawa, Ryunosuke Yoshino, Genki Kudo, Takatsugu Hirokawa. “Quantitative Estimation of Protein-Chemical Substructure Interaction with Inverse Mixed-Solvent Molecular Dynamics Simulation”, Chem-Bio Informatics Society(CBI) Annual Meeting 2023, 2023/10/23-26.
- 2023/09 An oral presentation will be given at an IIBMP2023 workshop.
- Keisuke Yanagisawa. “Further and Drastic Improvement of AlphaFold is Needed: Insights from Drug Design”, Informatics in Biology, Medicine, and Pharmacology 2023 (IIBMP2023): [WS-2] バイオインフォマティクスの8の問題, 2023/09/05.
2023 January-June
- 2023/06 Three oral presentations will be given at IPSJ SIG Bioinformatics and Genomics (SIGBIO).
- Jianan Li, Keisuke Yanagisawa, Masatake Sugita, Takuya Fujie, Masahito Ohue, Yutaka Akiyama. “CycPeptMPDB: Development of a Comprehensive Database of Membrane Permeability of Cyclic Peptides”, IPSJ SIG Technical Report, 2023-BIO-74(38): 1-8, 2023/07/01.
- Tomoya Saito, Keisuke Yanagisawa, Yutaka Akiyama. “フラグメント対の相対位置から検索可能な化合物立体配座データベースの構築”, IPSJ SIG Technical Report, 2023-BIO-74(37): 1-8, 2023/07/01.
- Ginga Watanabe, Keisuke Yanagisawa, Yutaka Akiyama. “標的RNAの高次構造予測に基づく低活性ASO候補配列の推測”, IPSJ SIG Technical Report, 2023-BIO-74(36): 1-8, 2023/07/01.
- 2023/03 A paper has been published.
- Jianan Li, Keisuke Yanagisawa, Masatake Sugita, Takuya Fujie, Masahito Ohue, Yutaka Akiyama. “CycPeptMPDB: A Comprehensive Database of Membrane Permeability of Cyclic Peptides”, Journal of Chemical Information and Modeling, 63: 2240-2250, 2023/03. DOI: 10.1021/acs.jcim.2c01573
- 2023/02 I got a JSPS KAKENHI (grant-in-aid for Scientific Research (B)) with two researchers.
- “Designing cyclic peptide with target protein selection based on mixed-solvent molecular dynamics simulation”, JSPS KAKENHI (Grant-in-Aid for Scientific Research (B)) 23H03495/23K28185, PI, 18 590 000 yen, 2023/04-2027/03.
2022 July-December
- 2022/10 two oral presentations and a poster presentation had been given at Chem-Bio Informatics Society(CBI) Annual Meeting 2022.
- Keisuke Yanagisawa, Rikuto Kubota, Yasushi Yoshikawa, Masahito Ohue, Yutaka Akiyama. “REstretto: An efficient protein-ligand docking tool based on a fragment reuse strategy”, Chem-Bio Informatics Society(CBI) Annual Meeting 2022, O2-1, 2022/10/25.
- Masatake Sugita, Takuya Fujie, Keisuke Yanagisawa, Masahito Ohue, Yutaka Akiyama. “Lipid composition is critical for accurate membrane permeability prediction of cyclic peptides by molecular dynamics simulations”, Chem-Bio Informatics Society(CBI) Annual Meeting 2022, O3-2, 2022/10/25.
- Genki Kudo, Keisuke Yanagisawa, Ryunosuke Yoshino, Takatsugu Hirokawa. “Amino Acid Preference Mapping on Protein-Protein Interaction Surface using Mixed-Solvent Molecular Dynamics”, Chem-Bio Informatics Society(CBI) Annual Meeting 2022, P02-04, 2022/10/25-27.
- 2022/09 A poster presentation will be given at the 60th Annual Meeting of the Biophysical Society of Japan.
- Keisuke Yanagisawa, Ryunosuke Yoshino, Genki Kudo, Takatsugu Hirokawa. “Inverse Mixed-Solvent Molecular Dynamics for Visualization of Amino Acid Residue Interaction Profile of Molecular Probes”, The 60th Annual Meeting of The Biophysical Society of Japan, 1Pos031, 2022/09/28.
- 2022/09 A paper has been published.
- Masatake Sugita, Takuya Fujie, Keisuke Yanagisawa, Masahito Ohue, Yutaka Akiyama. “Lipid composition is critical for accurate membrane permeability prediction of cyclic peptides by molecular dynamics simulations”, Journal of Chemical Information and Modeling, 62: 4549-4560, 2022/09. DOI: 10.1021/acs.jcim.2c00931
- 2022/08 A paper has been published.
- Keisuke Yanagisawa, Rikuto Kubota, Yasushi Yoshikawa, Masahito Ohue, Yutaka Akiyama. “Effective protein-ligand docking strategy via fragment reuse and a proof-of-concept implementation”, ACS Omega, 7: 30265-30274, 2022/08. DOI: 10.1021/acsomega.2c03470
2022 January-June
- 2022/06 An oral presentation will be given at IPSJ SIG Bioinformatics and Genomics (SIGBIO).
- Yudai Noso, Masatake Sugita, Takuya Fujie, Keisuke Yanagisawa, Masahito Ohue, Yutaka Akiyama. “分子動力学シミュレーション軌跡データからの環状ペプチドの膜透過性と相関が高い特徴量の抽出”, IPSJ SIG Technical Report, 2022-BIO-70(50): 1-8, 2022/06/29.
- 2022/06 An oral presentation will be given at the 22nd Annual Meeting of the Protein Science Society of Japan (PSSJ2022).
- Keisuke Yanagisawa, Ryunosuke Yoshino, Genki Kudo, Takatsugu Hirokawa. “Inverse Mixed-Solvent Molecular Dynamics for Visualization of Residue Interaction Profile of Molecular Probes”, The 22nd Annual Meeting of the Protein Science Society of Japan, O7-12, 2022/06/07.
- 2022/06 I got a grant for young researcher.
- “Recognition of protein surface where suitable for ligand binding with Inverse-MSMD simulations”, Grant for young researcher, School of Computing, Tokyo Institute of Technology, PI, 500 000 yen, 2022/06-2023/03.
- 2022/04 A paper has been published.
- Keisuke Yanagisawa, Ryunosuke Yoshino, Genki Kudo, Takatsugu Hirokawa. “Inverse Mixed-Solvent Molecular Dynamics for Visualization of the Residue Interaction Profile of Molecular Probes”, International Journal of Molecular Sciences, 23: 4749, 2022/04. DOI: 10.3390/ijms23094749
- 2022/03 Four oral presentation will be given at IPSJ SIG Bioinformatics and Genomics (SIGBIO).
- Yuki Tsushima, Keisuke Yanagisawa, Masahito Ohue, Yutaka Akiyama. “新たなデータセットによる長距離フラグメントリンキング手法の再評価”, IPSJ SIG Technical Report, 2022-BIO-69(16): 1-8, 2022/03/11.
- Masaya Inagaki, Keisuke Yanagisawa, Masahito Ohue, Yutaka Akiyama. “Database of Drug Candidates Represented by 3D Positional Relationships between Fragments”, IPSJ SIG Technical Report, 2022-BIO-69(15): 1-8, 2022/03/11.
- Mahiro Yamazaki, Kazuki Izawa, Ryo Hirata, Keisuke Yanagisawa, Masahito Ohue, Yutaka Akiyama. “Development of a Genome-wide Fast Short Nucleotide Sequence Search Method Considering Binding Energy”, IPSJ SIG Technical Report, 2022-BIO-69(8): 1-8, 2022/03/10.
- Shu Tamano, Kazuki Izawa, Keisuke Yanagisawa, Masahito Ohue, Yutaka Akiyama. “Proposal of Evaluation Off-target Effects Method in Gapmer ASO”, IPSJ SIG Technical Report, 2022-BIO-69(7): 1-7, 2022/03/10.
- 2022/02 A paper has been published.
- Kazuki Takabatake, Keisuke Yanagisawa, Yutaka Akiyama. “Solving Generalized Polyomino Puzzles Using the Ising Model”, Entropy, 24: 354, 2022/02. DOI: 10.3390/e24030354
News archive
Tools
EXPRORER (2021)
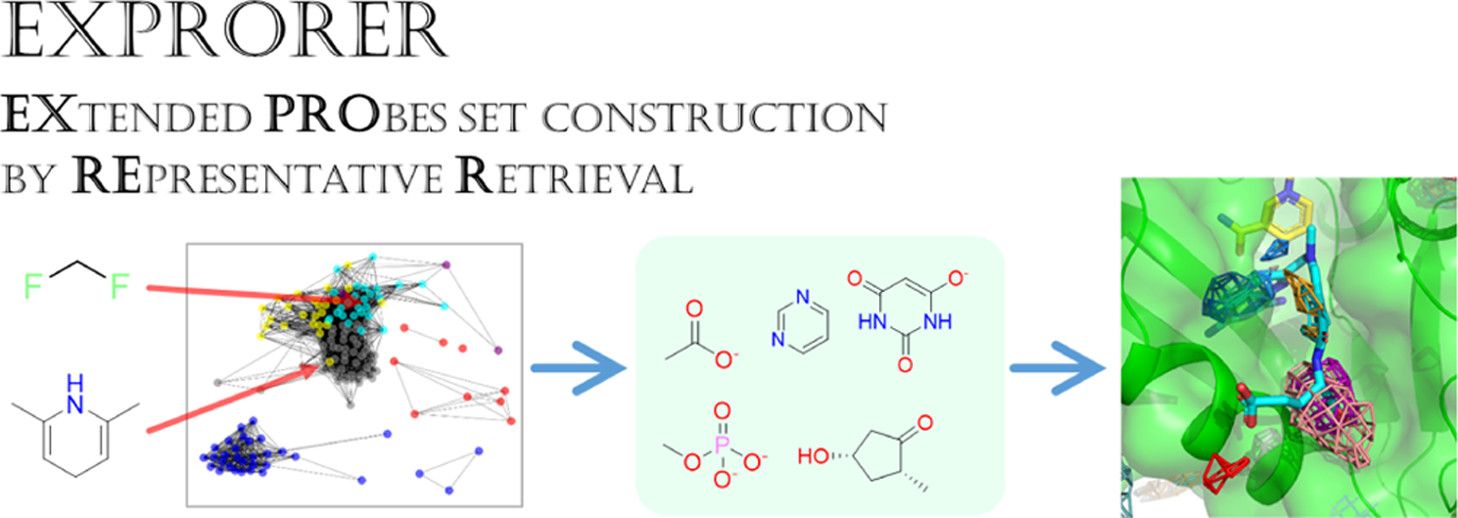
Rational cosolvent set construction with cosolvent MD simulation results
https://github.com/keisuke-yanagisawa/exprorer
Spresso (2017)

An ultrafast pre-screening method based on compound decomposition
